Research Article
Epigenetic Regulation of Hematological and Neurological Expressed 1(HN-1) at DU145 Brain Metastases Prostate Cancer Cell Line
Ceren Gonen-Korkmaz1*, Mehmet Zuhuri Arun1, Buket Reel1, Lokman Varıslı2 , Mazen Saeed Abdulaziz1 and Gokce YILDIRIM-BUHARALIOGLU1
1Department of Pharmacology, Ege University, Bornova, Izmir, Turkey
2Department of Bioengineering, Cancer Biology Laboratory, Ege University, Bornova, Izmir, Turkey
*Corresponding author: Ceren Gonen-Korkmaz, Department of Pharmacology, Ege University, Bornova, Izmir, Turkey
Published: 10 Jul, 2017
Cite this article as: Gonen-Korkmaz C, Arun MZ, Reel B,
Varıslı L, Abdulaziz MS, YILDIRIMBUHARALIOGLU
G. Epigenetic
Regulation of Hematological and
Neurological Expressed 1(HN-1) at
DU145 Brain Metastases Prostate
Cancer Cell Line. Clin Oncol. 2017; 2:
1314.
Abstract
Epigenetic investigates gene expression modifications not due to DNA sequence alterations. Gene
expression modifications appear with packing of DNA with variety of chromatin structures. The
most studied forms of epigenetic phenomenon are DNA methylation and histone modifications.
These pathways are connected with each other and reversible. DNA methylation reported genes:
36B4 (as control), HN1 primers were used and methylation status had been confirmed at DU145
cell line. Histone modifications which close the gene expression is histone deacetylation had been
reversed by Trichostatin A (TSA). It is reported that inhibition of methylation and deacetylation in
a serial way result with successful gene activation, performed with 5 Azacitidine and TSA incubation
at the project. In the recent years, epigenetic base had been shown for many diseases that research
and development for epigenetic disorders won successful treatment cases. Preclinical and clinic
levels of candidate drugs are on the way.
Keywords: HN1; DU145; TSA; 5-Azasitid
Abbreviations
ARE: Androgen Response Element; AZA: Azasitidin; HN1: Hematological and Neurological Expressed 1; TSA: Trikostatin A
Introduction
Cancer is a kind of disease which incidence has been increasing depending on the changing conditions of life and poses a grave threat to modern societies. Cancer which is characterized by uncontrolled cell growth and spread of abnormal cell structures to the organism is an epigenetic disease at the same level that it can be considered a genetic disease [1-3]. Therefore, cancer research has focused on epigenetic mechanisms that play critical role on carcinogenesis for the last decade. Within the scope of the project, a variety of gene expression changes were searched following demethylation [4] and inhibition of deacetylation [5] in prostate cancer cell line DU145 [6]. Also it was investigated whether there is a cross talk between the methylation and acetylation pathways after using the same demethylation agent with HDACI-Trikostatin A as a combination. Hematological and neurological expressed 1 (HN1) mRNA was first identified in the tissues of mouse embryos [7] and has also been detected in various other tissues. The gene is highly conserved in vertebrates [8] (although not in invertebrates), suggesting that HN1 has a conserved function in the development of the tissues in which is expressed. Furthermore, several studies have suggested that HN1 expression is associated with metastatic cancer progression [9] and neural development [10], as well as with nerve and retina regeneration [11]. Recently, Laughlin demonstrated that HN1 is involved in the malignant phenotypes of brain tumors [12] HN1 is a ubiquitously expressed, EGF-regulated gene. Expression of HN1 in prostate cell lines down-regulates PI3 K- dependent Akt activation [13]. HN1 is regulated by androgens through the putative androgen response elements (AREs) found in its promoter [14].
Materials and Methods
Cell culture
DU145 cells were cultured in DMEM-Ham’sF12 (GIBCO) with 5% fetal bovine serum, 1%
L-glutamine and 1 U/ml of each penicillin/ streptomycin. Cells were
incubated at 37°C and 5% CO2 in a humidified atmosphere.
RNA isolation
Qiagen R Neasy Mini Kit was used.
Epigenetic regulation
Histone deacetylase inhibition by Trichostatin A with 100 nM
for each plate and demethylation by 5-Azasitidin with 100 mg/ml for
each plate was used and overnight incubation were done.
Real-time PCR
Epigenetic genes primer couples for RT-PCR (Table 1) Real-time
PCR was performed using a Light Cycler® 480 (Roche Diagnostics)
instrument and Light Cycler 480 SYBR Green 1 Master (Roche
Diagnostics) kit. Briefly, reactions were performed in a 20 μl volume
with 5 pmol of each primer and 1 μl of cDNA template derived from
reverse-transcribed RNA of untreated control and incubated cells.
36B4 housekeeping gene was used as an endogenous control and
reference gene for relative quantifications. The same thermal profile
was optimized for all primers: a preincubation for 5 min at 950C
for 1 cycle, followed by 40 amplification cycles of denaturation at
95°C for 10 sec, a primer annealing at 64C for 20 sec, and a primer
extension at 72°C for 10 sec. Water was included as a no-template
control. Melting curves were derived aіer 40 cycles by a denaturation
step at 95°C for 10s, followed by annealing at 65°C for 15 sec, and a
temperature rise to 95°C with a heating rate of 0.1°C/s and continuous
fluorescence measurement. A final cooling was performed at 37°C for
30 sec. Melting curve analyses of each sample were performed using
Light Cycler 480 software version LCS480 (Roche Diagnostics). The
analysis step of relative quantification was a fully automated process
accomplished by the soіware, with the efficiency set at 2 and the
cDNA of untreated cells defined as calibrator. HN1 primers were
amplified using (10 pmol of each) primers which were designed using
Light Cycler Probe Design 6oіware 2 (Roche, Germany).
Statistical analysis
All the illustrated results represent one of at least three
independent experiments with similar outcomes.
Figure 1
Table 1
Results
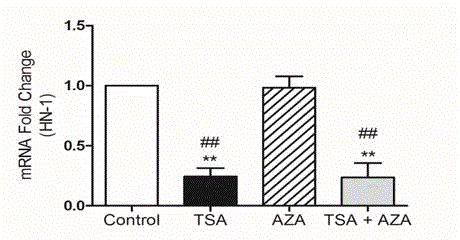
Epigenetic pathway was searched at DU145 cells. HDAC
Inhibition by Trichostatin A and demethylation by 5-Azasitidin had
been performed and HN1 specific primers were used at RT-PCR
(Figure 1).
**P ≤0.01 Versus Control, ##P ≤0.01 Versus AZA; Bonferroni Test
After Single Sided Anova Test.
TSA caused decrease versus control. (**P ≤0.01) and demethylation
with AZA respond as control and usage of both reagents decrease as
TSA (##P ≤0.01).
Discussion
Project was designed as to mimic severe carcinogenesis of prostate. DU145 is the brain metastases of prostate cancer reached to the target. Control group with closed histone showed HN1expression and inhibition of deacetylases with TSA, HN1 expression was decreased. As third group demethylation with 5-azasitidin also showed the expression level as control and it presents the homogeny expression distribution panel of HN1 [14]. Both reagents repeat the abundant role of TSA. The epigenetic regulation of HN1 had been shown for the first time in this report.
Acknowledgement
This study was supported by grants from The Scientific and Technological Research Council of Turkey (TUBITAK) to CGK (Grant no: 106S295) and EGE University Scientific Research Project no: 11ECZ017 to CGK.
References
- Baylin, Stephen B, Jones PA. A decade of exploring the cancer epigenome biological and translational implications. Nature Reviews Cancer. 2011; 11: 726-734.
- Chin SP, Dickinson JL, Holloway AF. Epigenetic regulation of prostate cancer Clin Epigenet. 2011; 2: 151-169.
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007; 128: 683-692.
- Jones PA, Taylor SM. Hemi ethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981; 9: 2933-2947.
- Korkmaz CG, Fronsdal K, Zhang Y, Lorenzo P, Saatcioglu F. Potentiating of androgen receptor transcriptional activity by inhibition of histone deacetylation - rescue of transcriptional compromised mutants. Journal of Endocrinology. 2004; 3: 377-389.
- Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145). Int J Cancer. 1978; 21: 274-281.
- Tang, W, Lai YH, Han XD, Wong PM, Peters LL, Chui DH. Murine Hn1 on chromosome 11 is expressed in hemopoietic and brain tissues. Mamm. Genome.1997; 8: 695-696.
- Zhou G, Wang J, Zhang Y, Zhong C, Ni J, Wang L, et al. Cloning, expression and sub cellular localization of HN1 and HN1L genes, as well as characterization of their orthologs, defining an evolutionarily conserved gene family. Gene. 2004; 331: 115-123.
- Huang GM, Ng WL, Farkas J, He L, Liang HA, Gordon D, et al. Prostate cancer expression profiling by cDNA sequencing analysis. Genomics. 1999; 59: 178-186.
- Zujovic V, Luo D, Baker HV, Lopez MC, Miller KR, Streit WJ, et al. The facial motor nucleus transcriptional program in response to peripheral nerve injury identifies Hn1 as a regeneration-associated gene. J Neurosci Res. 2005; 82: 581-591.
- Goto T, Hisatomi O, Kotoura M, Tokunagan F. Induced expression of hematopoietic- and neurologic-expressed sequence 1 in retinal pigment epithelial cells during newt retina regeneration. Exp Eye Res. 2006; 83: 972-980.
- Laughlin KM, Luo D, Liu C, Shaw G, Warrington Jr KH, Qiu J, et al. Hematopoietic- and neurologic-expressed sequence 1 expression in the murine GL261 and high-grade human gliomas. Pathol Oncol Res. 2009; 15: 437-444.
- Varışlı L, Gonen-Korkmaz C, Debeleç-Butuner B, Erbaykent-Tepedelen B, Mohammed H, Bogurcu N, et al. Ubiquitously expressed HN1 down-regulates Akt mediated GSK3beta signaling, and its knock down results with deregulated G2/M transition in prostate cells. DNA and Cell Biology. 2011; 30: 419-429.
- Varisli L, Gonen-Korkmaz C, Syed HM, Bogurcu N, Debelec-Butuner B, Erbaykent-Tepedelen B, et al. Androgen regulated HN1 leads proteosomal degradation of androgen receptor (AR) and negatively influences AR mediated transactivation in prostate cells. Mol Cell Endocrinol. 2012; 350: 107-117.