Editorial
NMR Metabolomics in Ionizing Radiation
Jian Zhi Hu*, Xiongjie Xiao and Mary Hu Y
Division of Earth and Biological Science, Pacific Northwest National Laboratory, USA
*Corresponding author: Jian Zhi Hu, Division of Earth and Biological Science, Pacific Northwest National Laboratory, WA 99354, USA
Published: 08 Sep, 2016
Cite this article as: Hu JZ, Xiao X, Mary Hu Y. NMR
Metabolomics in Ionizing Radiation. Clin
Oncol. 2016; 1: 1080.
Editorial
Ionizing radiation is an invisible threat that cannot be seen, touched or smelled and exists either
as particles or waves. Particle radiation can take the form of alpha, beta or neutrons, as well as high
energy space particle radiation such as high energy iron, carbon and proton radiation, etc [1]. Nonparticle
radiation includes gamma- and x-rays. Publically, there is a growing concern about the
adverse health effects due to ionizing radiation mainly because of the following facts. (a) The X-ray
diagnostic images are taken routinely on patients. Even though the overall dosage from a single
X-ray image such as a chest X-rays scan or a CT scan, also called X-ray computed tomography
(X-ray CT), is low, repeated usage can cause serious health consequences, in particular with the
possibility of developing cancer [2,3]. (b) Human space exploration has gone beyond moon and
is planning to send human to the orbit of Mars by the mid-2030s. And a landing on Mars will
follow. ("Obama PromisesRenewedSpace Program". The New York Times. Retrieved April 15, 2010).
Completely shield the high energy space radiation in outer space is a big challenging [4,5]. (c) The
impact of past nuclear disasters such as Chernobyl disaster (1986/4/26) and Fukushima Daiichi
nuclear disaster (2011/3/12) are long lasting, including leaving behand radiation contaminated sites
that are very difficult to clean [6,7]. And (d) Radiological hazards are likely to be employed by
terrorists via nuclear detonation, radiological dispersion devices, and covert placement/distribution
of radioactive substances [8]. The worst case scenario for a radiation incident would involve a
nuclear detonation-either from an improvised nuclear device or an actual warhead.
All cells can be damaged by ionizing radiation, but actively dividing cells are far more
radiosensitive than cells that are neither meiotically nor mitotically active. The most radiosensitive
cells in the human body include the bone marrow stem cells, gastrointestinal villi cells, and the
gametes in the ovaries and testes. Acute Radiation Syndrome (ARS) is an illness caused by partial
or whole-body exposure to high doses of ionizing radiation over a short period of time (usually
a few minutes or less). According to American military radiologists, the pathophysiology effects
dependence upon the irradiation doses are summarized in Table 1 [9]. Although the manifestations
of radiation injury vary depending on total absorbed radiation dose and the preexisting health of the
victim, it is clear from Table 1 that in most radiation scenarios, injury to the hematopoietic system
and GI tract are the main determinants of survival. If left untreated, a victim exposed to a total dose
of 3.5Gy (LD50 is about 4.0 Gy) and above is unlikely to survive.
The classical model of molecular injury involves immediate cellular damage following irradiation,
which can result in membrane and intracellular injury, i.e, inflammation, DNA single and double
strand break that subsequently turn on various genes and lead to cell proliferation, fibrosis, cancer
or cell death [10-12]. Significant investigations at molecular level have been done at the genetic
and protein levels by studying changes associated with DNA, RNA and proteins extracted from
cells and animal tissues using genomic [13,14] and proteomic [15,16] methods. Although expensive
and labor intensive, genomic and proteomic methods, may have potential as powerful tools for
studying different levels of the biological response to radiation-induced injury, including searching
for radiation specific molecular biomarkers. However, careful studies have generally shown a low
correlation between the pattern of gene expression and the pattern of protein expression [17,18].
Moreover, even in combination, genomic and proteomic methods still do not provide the range of
information needed for understanding integrated cellular function in a living system, since both
ignore the dynamic metabolic status of the whole organism.
It is well-known that alterations in DNA, RNA and protein are associated with changes in
metabolic profiles. Metabolites are chemical compounds that participate as reactants, intermediates,
or byproducts in a cellular metabolic pathway, and include carbon compounds with a molecular
weight typically in the range of 100-1000 Da. Radiation exposure will disturb the ratios and
concentrations of endogenous metabolites, either by direct chemical reaction or by binding to
key enzymes or nucleic acids that control metabolism. If these disturbances are of sufficient
magnitude, toxic effects will result. Therefore, metabolomics, defined
as a comprehensive and quantitative analysis of all metabolites in a
biological system [19-21], will be an important new systems biology
tool for elucidating the molecular mechanisms of radiation.
Metabolomics is a new technique and has only been recently
applied in the field of radiation, emerging as a field of great
significance for both translational and basic research [22-25]. Unlike
approaches in which biomolecules/metabolites are selected and
analyzed one or a few at a time, metabolomics focuses on broad
identification and analysis of multiple metabolites simultaneously.
The state of metabolome cumulatively reflects the stages of gene
expression, protein expression, and the cellular environment as well
as multidirectional interactions among these elements. Metabolomic
information is complementary, yet distinct, from that generated
by genomic and proteomic approaches. Moreover, metabolic
changes are among the earliest cellular responses to environmental
or physiological changes. It is well-known that there are estimated
30,000-40,000 genes (genome) associated with DNA, more than
100,000 transcripts (transcriptome) associated with RNA, and more
than 1,000,000 proteins (proteome) yet there are only approximately
5000 metabolites (metabolome) in human cells [26,27]. It is clear that
complexity is greatly simplified with metabolomics which, although
in its infancy, has already proven capable of detecting and diagnosing
a disease and evaluating the efficacy of therapy in an early stage
[22,23,25,28]. Therefore, it is highly likely that metabolomics will
provide valuable new information about the impact of radiation on
human health.
Nuclear Magnetic Resonance (NMR) spectroscopy is a
quantitative, non-destructive method that requires no or minimal
sample preparation, and is one of the leading analytical tools for
metabonomic research [19,29-33]. Unlike mass spectrometry
based methods, where the peak intensity depends on the efficiency
of ionization of the molecules that are different for different
types of molecules and the ion suppression issues when multiple
species coelute, the peak intensity in an NMR spectrum is directly
proportional to the number or concentration of molecules. The
easy quantification associated with NMR is a big advantage over
other techniques. 1H NMR is especially attractive because protons
are present in virtually all metabolites and its NMR sensitivity is
high, enabling the simultaneous identification and monitoring of
a wide range of low molecular weight metabolites, thus providing
a biochemical fingerprint of an organism “without prejudice”. It
is expected that NMR metabolomics will play an important role in
understanding the damage at molecular level by ionizing radiation as
have demonstrated recently by us [34,35].
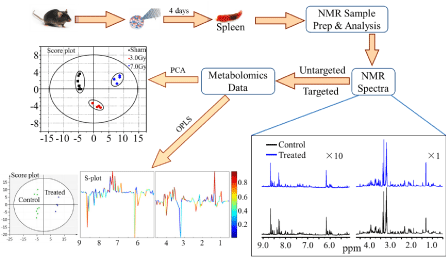
Figure 1 shows an example [35] of applying 1H NMR
metabolomics to study the changes in metabolic profile in the
spleen of C57BL/6 mouse after 4 days whole body exposure to 3.0
Gy and 7.8 Gy gamma radiations. As an integrated part of NMR
metabolomics, principal component analysis (PCA) [36], an
unsupervised statistical method, and orthogonal projection to latent
structures analysis (OPLS) [37], a supervised statistical method, are
employed for classification and identification of potential biomarkers
associated with gamma irradiation. The results from the PCA and
OPLS analysis have shown [35] that the exposed groups can be well
separated from the control group. Leucine, 2-aminobutyrate, valine,
lactate, arginine, glutathione, 2-oxoglutarate, creatine, tyrosine,
phenylalanine, π-methylhistidine, taurine, myo-inositol, glycerol and
uracil are significantly elevated while ADP is decreased significantly.
These significantly changed metabolites are associated with multiple
metabolic pathways and may be considered as potential biomarkers
in the spleen exposed to gamma irradiation.
Table 1
Figure 1
Figure 1
Example of applying 1H NMR metabolomics to study the changes in metabolic profile in the spleen of C57BL/6 mouse after 4 days whole body exposure
to 3.0 Gy and 7.8 Gy gamma radiations.
Acknowledgement
The preparation of this editorial note was supported by the National Institute of Environmental Health Sciences of the National Institute of Health (NIH) under Award Number R01ES022176, and was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE's Office of Biological and Environmental Research, and located at Pacific Northwest National Laboratory (PNNL). PNNL is a multi-program national laboratory operated for the DOE by Battelle Memorial Institute under Contract DE-AC06-76RLO 1830.
References
- Lloyd SAJ, Bandstra ER, Travis ND, Nelson GA, Bourland JD, Pecaut MJ, et al. Spaceflight-relevant types of ionizing radiation and cortical bone: Potential LET effect? Advances in Space Research. 2008; 42: 1889-1897.
- Brenner DJ, Hall EJ. Current concepts-Computed tomography-An increasing source of radiation exposure. New Engl J Med. 2007; 357: 2277-2284.
- de Gonzalez AB, Mahesh M, Kim KP, Bhargavan M, Lewis R, Mettler F, et al. Projected Cancer Risks From Computed Tomographic Scans Performed in the United States in 2007. Arch Intern Med. 2009; 169: 2071-2077.
- Townsend LW. Critical analysis of active shielding methods for space radiation protection. 2005 IEEE Aerospace Conference. 2005.
- Battiston R, Burger WJ, Calvelli V, Musenich R, Choutko V, Datskov VI, et al. Active Radiation Shield for Space Exploration Missions (ARSSEM). 2012.
- Bouville A. The Chernobyl Accident. Radiat Prot Dosim. 1995; 60: 287-293.
- Cho HS, Woo TH. project strategy for clean-up of sedimentary radioactive material in fukushima bay areas using snake-like robotics. Nucl Technol Radiat Prot. 2015; 30: 318-323.
- Manthous CA, Jackson WL. The 9-11 Commission's invitation to imagine: A pathophysiology-based approach to critical care of nuclear explosion victims. Crit Care Med. 2007; 35: 716-723.
- Anno GH, Baum SJ, Withers HR, Young RW. Symptomatology of Acute Radiation Effects in Humans after Exposure to Doses of 0.5-30 Gy. Health Phys. 1989; 56: 821-838.
- Jonathan EC, Bernhard EJ, McKenna WG. How does radiation kill cells? Current opinion in chemical biology. 1999; 3: 77-83.
- Somosy Z. Radiation response of cell organelles. Micron. 2000; 31: 165-181.
- Kim KC, Lee IK, Kang KA, Kim BJ, Kim D, Moon JY, et al. Empetrum nigrum var. japonicum Extract Suppresses gamma-Ray Radiation-Induced Cell Damage via Inhibition of Oxidative Stress. Am J Chin Med. 2011; 39: 161-170.
- Szumiel I. Ionizing radiation-induced oxidative stress, epigenetic changes and genomic instability: The pivotal role of mitochondria. Int J Radiat Biol. 2015; 91: 1-12.
- Nguyen V, Panyutin IV, Panyutin IG, Neumann RD. A Genomic Study of DNA Alteration Events Caused by Ionizing Radiation in Human Embryonic Stem Cells via Next-Generation Sequencing. Stem Cells Int. 2016: 7.
- Bakshi MV, Barjaktarovic Z, Azimzadeh O, Kempf SJ, Merl J, Hauck SM, et al. Long-term effects of acute low-dose ionizing radiation on the neonatal mouse heart: a proteomic study. Radiat Environ Bioph. 2013; 52: 451-461.
- Vieira HGS, Grynberg P, Bitar M, Pires SD, Hilario HO, Macedo AM, et al. Proteomic Analysis of Trypanosoma cruzi Response to Ionizing Radiation Stress. Plos One. 2014; 9: 16.
- Gry M, Rimini R, Stromberg S, Asplund A, Ponten F, Uhlen M, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. Bmc Genomics. 2009; 10.
- Nakaminami K, Matsui A, Nakagami H, Minami A, Nomura Y, Tanaka M, et al. Analysis of Differential Expression Patterns of mRNA and Protein During Cold-acclimation and De-acclimation in Arabidopsis. Mol Cell Proteomics. 2014; 13: 3602-3611.
- Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999; 29:1181-1189.
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000; 18: 1157-1161.
- Nicholson JK, Lindon JC. Systems biology-Metabonomics. Nature. 2008; 455: 1054-1056.
- Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Gonzalez FJ, et al. Radiation metabolomics. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008; 170: 1-14.
- Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ Jr, Gonzalez FJ, et al. Radiation Metabolomics. 2. Dose- and Time-Dependent Urinary Excretion of Deaminated Purines and Pyrimidines after Sublethal Gamma-Radiation Exposure in Mice. Radiat Res. 2009; 172: 42-57.
- Cheema AK, Pathak R, Zandkarimi F, Kaur P, Alkhalil L, Singh R, et al. Liver Metabolomics Reveals Increased Oxidative Stress and Fibrogenic Potential in Gfrp Transgenic Mice in Response to Ionizing Radiation. J Proteome Res. 2014; 13: 3065-3074.
- Laiakis EC, Trani D, Moon BH, Strawn SJ, Fornace AJ. Metabolomic Profiling of Urine Samples from Mice Exposed to Protons Reveals Radiation Quality and Dose Specific Differences. Radiat Res. 2015; 183: 382-390.
- Nørregaard Jensen O. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Current Opinion in Chemical Biology. 2004; 8: 33-41.
- Horgan RP, Kenny LC. ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. The Obstetrician & Gynaecologist. 2011; 13: 189-195.
- Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace AJ. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011; 87: 802-823.
- Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HWL, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics vol 8, pg 1439, 2002. Nat Med. 2003; 9: 477.
- Coen M, Holmes E, Lindon JC, Nicholson JK. NMR-based metabolic profiling and metabonomic approaches to problems in molecular toxicology. Chem Res Toxicol. 2008; 21: 9-27.
- Jiang LM, Huang J, Wang YL, Tang HR. Eliminating the dication-induced intersample chemical-shift variations for NMR-based biofluid metabonomic analysis. Analyst. 2012; 137: 4209-4219.
- Schicho R, Shaykhutdinov R, Ngo J, Nazyrova A, Schneider C, Panaccione R, et al. Quantitative metabolomic profiling of serum, plasma, and urine by (1) H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J Proteome Res. 2012; 11: 3344-3357.
- Yang YX, Wang LJ, Wang SM, Liang SW, Chen A, Tang HR, et al. Study of metabonomic profiles of human esophageal carcinoma by use of high-resolution magic-angle spinning H-1 NMR spectroscopy and multivariate data analysis. Anal Bioanal Chem. 2013; 405: 3381-3389.
- Zhang Q, Hu JZ, Rommereim DN, Murphy MK, Phipps RP, Huso DL, et al. Application of High-Resolution H-1 MAS NMR Spectroscopy to the Analysis of Intact Bones from Mice Exposed to Gamma Radiation. Radiat Res. 2009; 172: 607-616.
- Xiao X, Hu M, Liu M, Hu J. 1H NMR Metabolomics Study of Spleen from C57BL/6 Mice Exposed to Gamma Radiation. Metabolomics (Open Access). 2016; 6: 1-11.
- Hotelling H. Analysis of a complex of statistical variables into principal components. J Educ Psychol. 1933; 24: 417-441.
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemometr. 2002; 16: 119-128.